


Hello! 👋 I am Co-Founder, CTO and Chief Scientist @ molecule.one, where we build a highly automated chemistry platform for synthesis and autonomous scientific discovery. I am also Area Chair for ICLR 2026 and Venture Advisor at Expeditions Fund.
I am passionate about understanding natural and artificial learning systems, from deep neural networks to the scientific process and economy. I believe that we should look for inspiration in such natural processes to build neural networks capable of continual learning, creativity and high sample efficiency.
I am now applying this optimization lens to autonomous discovery of new chemistry at molecule.one.
Previously, during my PhD, I studied the foundations of gradient-based training and co-authored work showing how optimization dynamics drives neural networks into high-curvature chaotic regimes, helping explain the role of learning rate in generalization and training stability (ICLR 2020 spotlight; also covered in Understanding Deep Learning, Prince, MIT Press).
I joined molecule.one as a co-founder to build and lead the development of MARIA, a first-of-its-kind high-throughput chemistry platform controlled by state-of-the-art AI. We named the platform after Maria Skłodowska-Curie, a double Nobel laureate to reflect the goal to both automate chemistry and push the boundaries of science. We applied the platform across drug discovery and chemical discovery with multiple partners, notably enabling synthesis of thousands of highly novel molecules per week.
Before that, I did postdoc at New York University with Kyunghyun Cho and Krzysztof Geras, and before that PhD at Jagiellonian University. My PhD was focused on the foundations of deep neural networks. I was fortunate to collaborate and co-author highly cited papers on the topic with MILA (with Yoshua Bengio), University of Edinburgh (with my co-supervisor Amos Storkey) and Google Research.
I contribute to the scientific community as an Action Editor for TMLR and an area chair for ICLR 2026 (before that NeurIPS 2020-25, ICML 2020-22, ICLR 2020-25).






For a full list please see my Google Scholar profile.
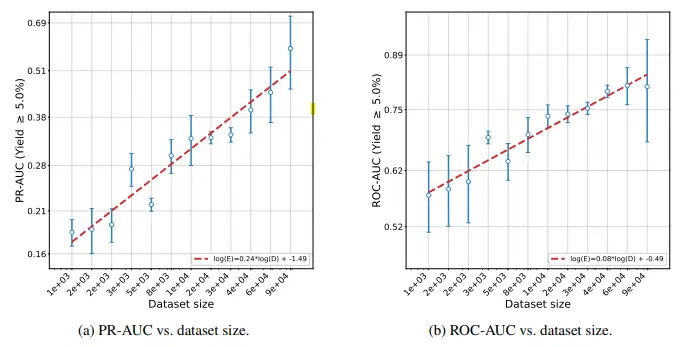
M. Sadowski, L. Sztukiewicz, M. Wyrzykowska, [...], S. Jastrzebski
NeurIPS 2025 Workshop AI4Science
paper
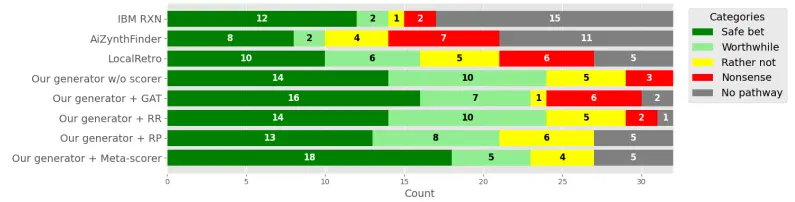
M. Sadowski, [...], S. Jastrzebski
NeurIPS 2025 Workshop AI4Science
paper
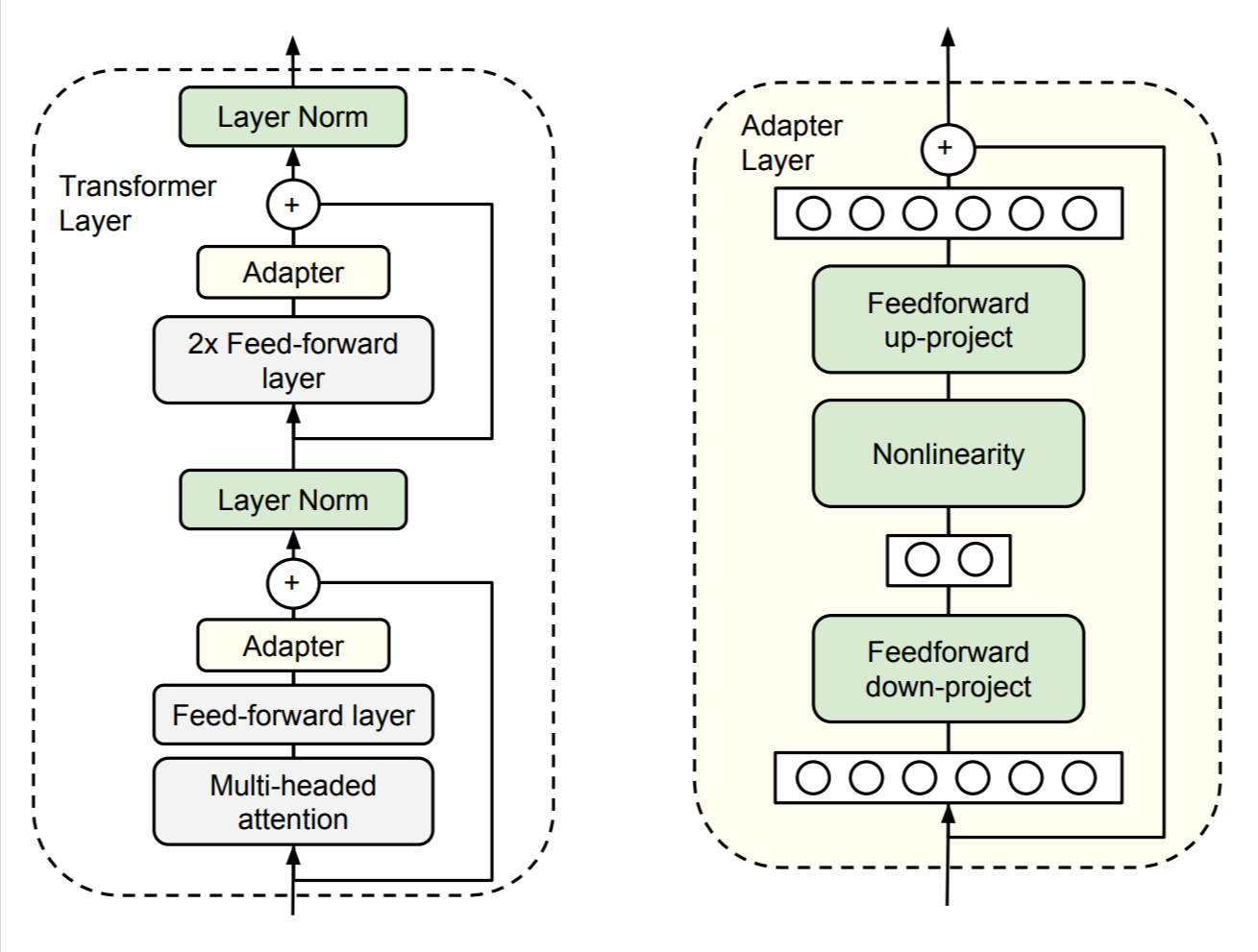
S. Houlsby, A. Giurgiu, S. Jastrzebski, B. Morrone, Q. de Laroussilhe, A. Gesmundo, M. Attariyan, S. Gelly
International Conference on Machine Learning 2019
paper
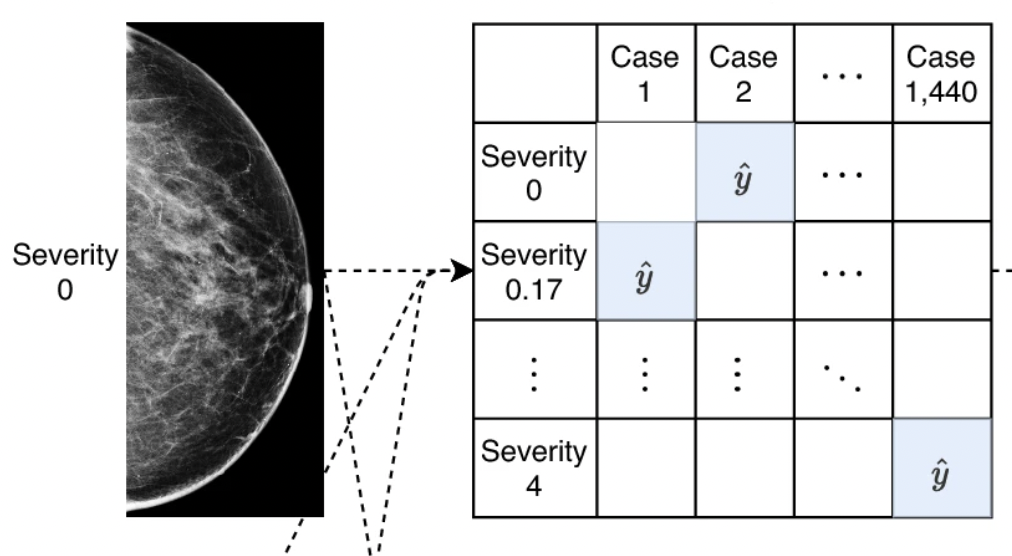
T. Makino, S. Jastrzebski, Witold Oleszkiewicz, [...], Kyunghyun Cho, Krzysztof J Geras
Nature Scientific Reports 2022
paper
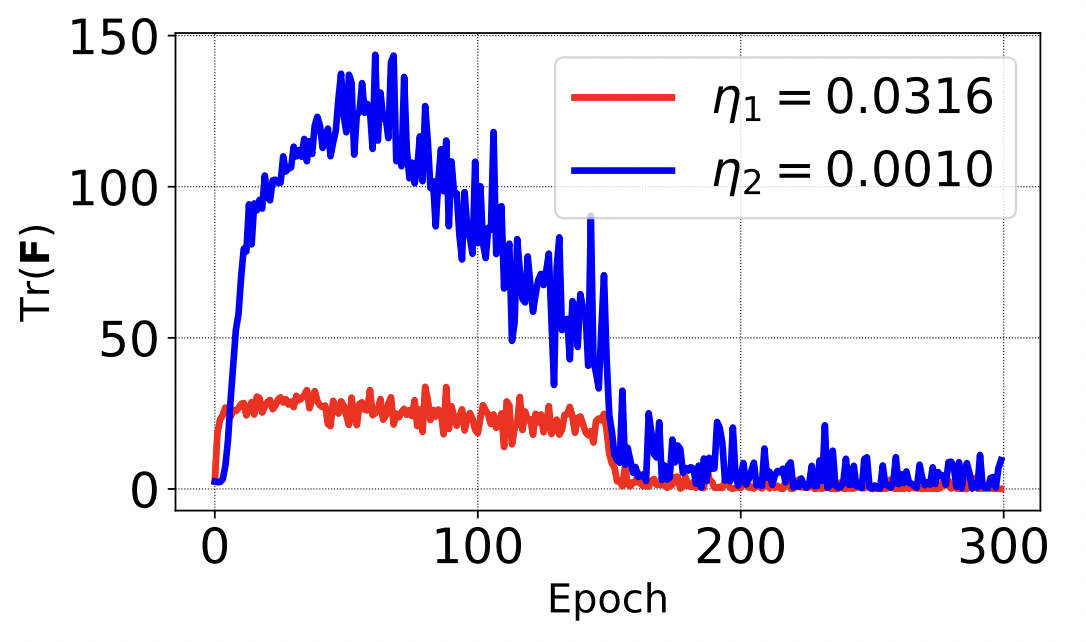
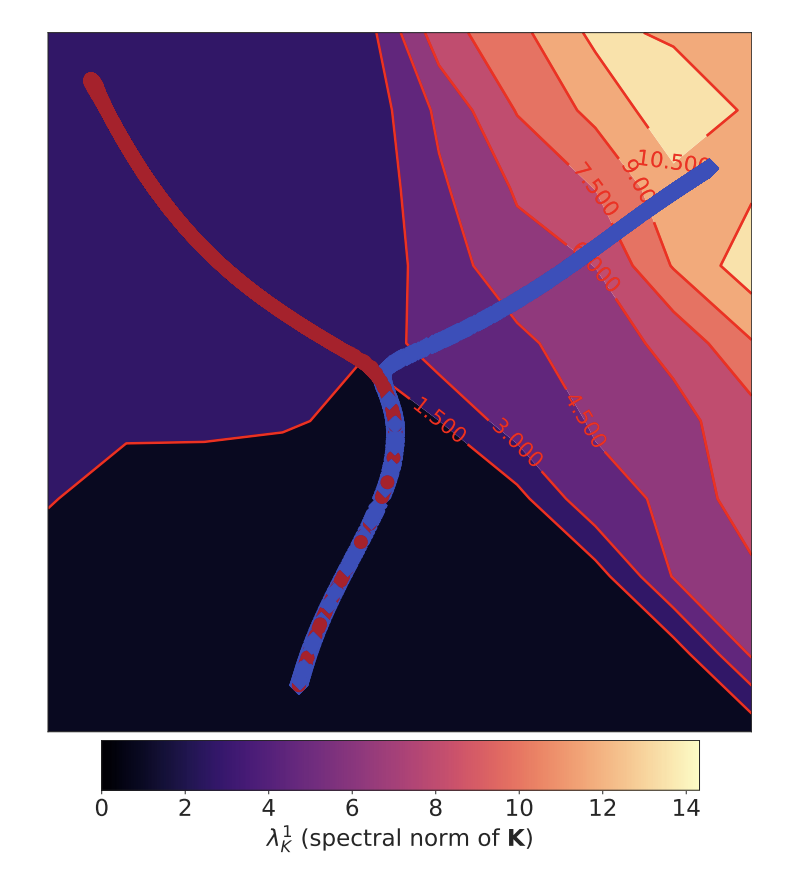
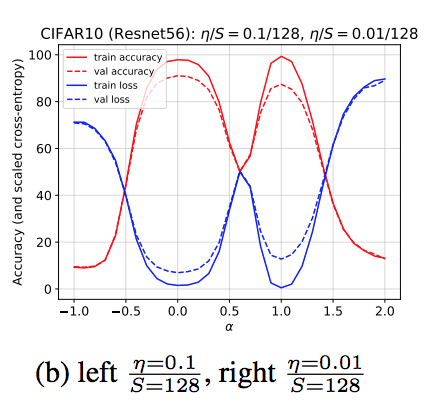
S. Jastrzębski*, Z. Kenton*, D. Arpit, N. Ballas, A. Fischer, Y. Bengio, A. Storkey
International Conference on Artificial Neural Networks 2018 (oral), International Conference on Learning Representations 2018 (workshop)
paper